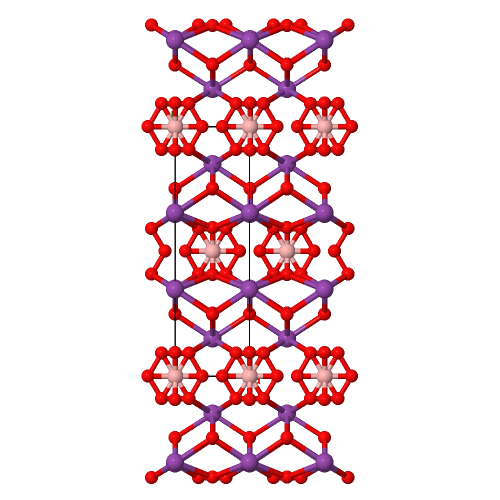
When the difference between incommensurate and commensurate structures becomes elusive: the case of Bi4BO7X (X = Cl, Br) oxyhalides
Authors:Volkov, Sergey; Arsentev, Maxim; Ugolkov, Valery; Povolotskiy, Alexey; Savchenko, Yevgeny; Krzhizhanovskaya, Maria; Bubnova, Rimma; Aksenov, Sergey
Journal:Journal of Applied Crystallography 56 589-596 (2023)
DOI:https://doi.org/10.1107/S1600576723002261
B-IncStrDB ID: aZTkbw2u06w Entry date: 2025-09-22 Last revision: 2025-09-22
I
Chemical data
Structural Formula Sum: B Bi4 Cl O7 [ Help ]
Formula weight: 994.2 Da [ Help ]
Absolute configuration: . [ Help ]
Crystallographic data and experimental details
Crystal system: orthorhombic [ Help ]
Superspace group name: Immm(00γ)000 [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,-x2,x3,x4 |
| 3 | -x1,x2,-x3,-x4 |
| 4 | x1,-x2,-x3,-x4 |
| 5 | -x1,-x2,-x3,-x4 |
| 6 | x1,x2,-x3,-x4 |
| 7 | x1,-x2,x3,x4 |
| 8 | -x1,x2,x3,x4 |
| 9 | x1+1/2,x2+1/2,x3+1/2,x4 |
| 10 | -x1+1/2,-x2+1/2,x3+1/2,x4 |
| 11 | -x1+1/2,x2+1/2,-x3+1/2,-x4 |
| 12 | x1+1/2,-x2+1/2,-x3+1/2,-x4 |
| 13 | -x1+1/2,-x2+1/2,-x3+1/2,-x4 |
| 14 | x1+1/2,x2+1/2,-x3+1/2,-x4 |
| 15 | x1+1/2,-x2+1/2,x3+1/2,x4 |
| 16 | -x1+1/2,x2+1/2,x3+1/2,x4 |
a: 3.92770(10) Å [ Help ]
b: 13.0981(5) Å [ Help ]
c: 3.88080(10) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 199.649(11) Å3 [ Help ]
Modulation dimension: 1 [ Help ]
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.000000 | 0.000000 | 0.248400 |
Z: 1 [ Help ]
Cell determination reflection Nb.: 1154 [ Help ]
θ(min) for cell determination: 5.39 ° [ Help ]
θ(max) for cell determination: 33.55 ° [ Help ]
Cell measurement temperature: 296.9(7) K [ Help ]
μ: 88.211 mm-1 [ Help ]
Absorption correction type: gaussian [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.41.93a (Rigaku Oxford Diffraction, 2020) Numerical absorption correction based on gaussian integration over a multifaceted crystal model Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Minimum transmission factor: 0.045 [ Help ]
Maximum transmission factor: 0.248 [ Help ]
Refinement details
Total nb. of reflections: 1489 [ Help ]
Nb. of observed reflections: 982 [ Help ]
Intense reflections threshold: I>3σ(I) [ Help ]
Refinement based on: F [ Help ]
R(obs): 0.0664 [ Help ]
wR(obs): 0.0880 [ Help ]
R(all): 0.0848 [ Help ]
wR(all): 0.0901 [ Help ]
S(all): 4.08 [ Help ]
S(obs): 4.96 [ Help ]
Nb. of reflections: 1489 [ Help ]
Nb. of parameters: 51 [ Help ]
Number of restraints: 0 [ Help ]
Number of constraints: 10 [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/(σ2(F)+0.0001F2) [ Help ]
Δ/σ(max): 0.0357 [ Help ]
Δ/σ(mean): 0.0071 [ Help ]
Δρ(max): 3.43 e_Å-3 [ Help ]
Δρ(min): -12.85 e_Å-3 [ Help ]
Extinction method: none [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | ADP type | Uiso/equiv | Symmetry multiplicity | Occupancy | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bi | Bi | 0.5 | 0.15371(3) | 0.5 | Uani | 0.02037(19) | 4 | 1 | d | . | . | . |
| Cl | Cl | 0 | 0 | 0 | Uani | 0.044(2) | 2 | 0.5 | d | . | . | . |
| B | B | 0 | 0 | 0 | Uani | 0.044(2) | 2 | 0.5 | d | . | . | . |
| O1 | O | 0 | 0.2474(7) | 0.5 | Uani | 0.019(2) | 4 | 1 | d | . | . | . |
| O2 | O | 0 | -0.0944(14) | 0 | Uani | 0.021(5) | 4 | 0.25 | d | . | . | . |
| O3 | O | -0.184(8) | -0.092(3) | 0 | Uani | 0.038 | 8 | 0.125 | d | . | . | . |
| O4 | O | -0.363 | 0 | 0 | Uani | 0.131 | 4 | 0.25 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | Atom site symbol | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|---|
| Bi | Bi | 0.0254(4) | 0.0179(3) | 0.0178(3) | 0 | 0 | 0 |
| Cl | Cl | 0.063(5) | 0.028(4) | 0.041(4) | 0 | 0 | 0 |
| B | B | 0.063(5) | 0.028(4) | 0.041(4) | 0 | 0 | 0 |
| O1 | O | 0.018(4) | 0.025(5) | 0.014(3) | 0 | 0 | 0 |
| O2 | O | 0.018(8) | 0.014(9) | 0.031(9) | 0 | 0 | 0 |
| O3 | O | 0.037995 | 0.037995 | 0.037995 | 0 | 0 | 0 |
| O4 | O | 0.130968 | 0.130968 | 0.130968 | 0 | 0 | 0 |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_1 |
|---|---|
| 1 | 1 |
| 2 | 2 |
| 3 | 3 |
Occupation crenel coefficients: (Show/hide table) [ Help ]
| Atom site label | Center (x0) | Width |
|---|---|---|
| Cl | 0.5 | 0.5 |
| B | 0 | 0.5 |
| O2 | 0 | 0.5 |
| O3 | 0 | 0.5 |
| O4 | 0 | 0.5 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Bix1 | Bi | x | 1 |
| Biy1 | Bi | y | 1 |
| Biz1 | Bi | z | 1 |
| Bix2 | Bi | x | 2 |
| Biy2 | Bi | y | 2 |
| Biz2 | Bi | z | 2 |
| Bix3 | Bi | x | 3 |
| Biy3 | Bi | y | 3 |
| Biz3 | Bi | z | 3 |
| O1x1 | O1 | x | 1 |
| O1y1 | O1 | y | 1 |
| O1z1 | O1 | z | 1 |
| O1x2 | O1 | x | 2 |
| O1y2 | O1 | y | 2 |
| O1z2 | O1 | z | 2 |
| O1x3 | O1 | x | 3 |
| O1y3 | O1 | y | 3 |
| O1z3 | O1 | z | 3 |
| O2x1 | O2 | x | 1 |
| O2y1 | O2 | y | 1 |
| O2z1 | O2 | z | 1 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Bix1 | 0 | 0 |
| Biy1 | -0.00425(4) | 0 |
| Biz1 | 0 | -0.02719(15) |
| Bix2 | 0 | 0 |
| Biy2 | -0.00062(7) | 0 |
| Biz2 | 0 | -0.00265(11) |
| Bix3 | 0 | 0 |
| Biy3 | 0.00024(5) | 0 |
| Biz3 | 0 | 0.00893(9) |
| O1x1 | 0 | 0 |
| O1y1 | 0.0039(6) | 0 |
| O1z1 | 0 | 0.045(2) |
| O1x2 | 0 | 0 |
| O1y2 | -0.0057(10) | 0 |
| O1z2 | 0 | -0.0124(18) |
| O1x3 | 0 | 0 |
| O1y3 | 0.0034(7) | 0 |
| O1z3 | 0 | 0.0053(14) |
| O2x1 | 0 | 0 |
| O2y1 | 0 | 0 |
| O2z1 | 0 | 0.155(7) |
Displacive (translational) Legendre coefficients: (Show/hide table) [ Help ]
| Atom site label | Displacement axis | Polynomial order | Polynomial coeff. |
|---|---|---|---|
| Cl | x | 1 | 0 |
| Cl | y | 1 | 0 |
| Cl | z | 1 | 0.009(3) |
| Cl | x | 2 | 0 |
| Cl | y | 2 | 0 |
| Cl | z | 2 | 0 |
Definition of the ADP Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Tensor element | Wave vector code |
|---|---|---|---|
| BiU111 | Bi | U11 | 1 |
| BiU221 | Bi | U22 | 1 |
| BiU331 | Bi | U33 | 1 |
| BiU121 | Bi | U12 | 1 |
| BiU131 | Bi | U13 | 1 |
| BiU231 | Bi | U23 | 1 |
| BiU112 | Bi | U11 | 2 |
| BiU222 | Bi | U22 | 2 |
| BiU332 | Bi | U33 | 2 |
| BiU122 | Bi | U12 | 2 |
| BiU132 | Bi | U13 | 2 |
| BiU232 | Bi | U23 | 2 |
| BiU113 | Bi | U11 | 3 |
| BiU223 | Bi | U22 | 3 |
| BiU333 | Bi | U33 | 3 |
| BiU123 | Bi | U12 | 3 |
| BiU133 | Bi | U13 | 3 |
| BiU233 | Bi | U23 | 3 |
| O1U111 | O1 | U11 | 1 |
| O1U221 | O1 | U22 | 1 |
| O1U331 | O1 | U33 | 1 |
| O1U121 | O1 | U12 | 1 |
| O1U131 | O1 | U13 | 1 |
| O1U231 | O1 | U23 | 1 |
ADP Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| BiU111 | 0.0119(2) | 0 |
| BiU221 | 0.0076(2) | 0 |
| BiU331 | 0.0113(2) | 0 |
| BiU121 | 0 | 0 |
| BiU131 | 0 | 0 |
| BiU231 | 0 | 0.00306(15) |
| BiU112 | -0.0035(3) | 0 |
| BiU222 | 0.0011(4) | 0 |
| BiU332 | 0.0054(2) | 0 |
| BiU122 | 0 | 0 |
| BiU132 | 0 | 0 |
| BiU232 | 0 | -0.0009(3) |
| BiU113 | 0.0028(2) | 0 |
| BiU223 | 0.0020(2) | 0 |
| BiU333 | 0.00152(16) | 0 |
| BiU123 | 0 | 0 |
| BiU133 | 0 | 0 |
| BiU233 | 0 | -0.0017(2) |
| O1U111 | 0.009(3) | 0 |
| O1U221 | 0.008(4) | 0 |
| O1U331 | -0.009(3) | 0 |
| O1U121 | 0 | 0 |
| O1U131 | 0 | 0 |
| O1U231 | 0 | -0.007(3) |
ADP Legendre coefficients: (Show/hide table) [ Help ]
| Atom site label | Tensor element | Polynomial order | Polynomial coeff. |
|---|---|---|---|
| Cl | U11 | 1 | 0 |
| Cl | U22 | 1 | 0 |
| Cl | U33 | 1 | 0 |
| Cl | U12 | 1 | 0 |
| Cl | U13 | 1 | 0 |
| Cl | U23 | 1 | 0 |
| Cl | U11 | 2 | 0.033(13) |
| Cl | U22 | 2 | -0.020(10) |
| Cl | U33 | 2 | 0.031(7) |
| Cl | U12 | 2 | 0 |
| Cl | U13 | 2 | 0 |
| Cl | U23 | 2 | 0 |
II
Chemical data
Structural Formula Sum: Bi4 B O7 Br [ Help ]
Formula weight: 1038.6 Da [ Help ]
Absolute configuration: . [ Help ]
Crystallographic data and experimental details
Crystal system: orthorhombic [ Help ]
Superspace group name: Immm(00γ)000 [ Help ]
Symmetry operations of the superspace group: (Show/hide table) [ Help ]
| Operation code | Operation in algebraic form |
|---|---|
| 1 | x1,x2,x3,x4 |
| 2 | -x1,-x2,x3,x4 |
| 3 | -x1,x2,-x3,-x4 |
| 4 | x1,-x2,-x3,-x4 |
| 5 | -x1,-x2,-x3,-x4 |
| 6 | x1,x2,-x3,-x4 |
| 7 | x1,-x2,x3,x4 |
| 8 | -x1,x2,x3,x4 |
| 9 | x1+1/2,x2+1/2,x3+1/2,x4 |
| 10 | -x1+1/2,-x2+1/2,x3+1/2,x4 |
| 11 | -x1+1/2,x2+1/2,-x3+1/2,-x4 |
| 12 | x1+1/2,-x2+1/2,-x3+1/2,-x4 |
| 13 | -x1+1/2,-x2+1/2,-x3+1/2,-x4 |
| 14 | x1+1/2,x2+1/2,-x3+1/2,-x4 |
| 15 | x1+1/2,-x2+1/2,x3+1/2,x4 |
| 16 | -x1+1/2,x2+1/2,x3+1/2,x4 |
a: 3.9345(2) Å [ Help ]
b: 13.2499(5) Å [ Help ]
c: 3.9182(2) Å [ Help ]
α: 90 ° [ Help ]
β: 90 ° [ Help ]
γ: 90 ° [ Help ]
Volume: 204.263(17) Å3 [ Help ]
Modulation dimension: 1 [ Help ]
Measured independent wave vectors: (Show/hide table) [ Help ]
| Wave vector id | q_x | q_y | q_z |
|---|---|---|---|
| 1 | 0.000000 | 0.000000 | 0.245700 |
Z: 1 [ Help ]
Cell determination reflection Nb.: 1217 [ Help ]
θ(min) for cell determination: 5.39 ° [ Help ]
θ(max) for cell determination: 33.63 ° [ Help ]
Cell measurement temperature: 297.7(9) K [ Help ]
μ: 90.782 mm-1 [ Help ]
Absorption correction type: gaussian [ Help ]
Absorption correction remarks: CrysAlisPro 1.171.41.93a (Rigaku Oxford Diffraction, 2020) Spherical absorption correction using equivalent radius and absorption coefficient. Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. [ Help ]
Minimum transmission factor: 0.753 [ Help ]
Maximum transmission factor: 0.76 [ Help ]
Refinement details
Total nb. of reflections: 1333 [ Help ]
Nb. of observed reflections: 751 [ Help ]
Intense reflections threshold: I>3σ(I) [ Help ]
Refinement based on: F [ Help ]
R(obs): 0.0476 [ Help ]
wR(obs): 0.0551 [ Help ]
R(all): 0.0860 [ Help ]
wR(all): 0.0634 [ Help ]
S(all): 2.22 [ Help ]
S(obs): 2.60 [ Help ]
Nb. of reflections: 1333 [ Help ]
Nb. of parameters: 32 [ Help ]
Number of restraints: 0 [ Help ]
Number of constraints: 13 [ Help ]
Weighting scheme: sigma [ Help ]
Weighting scheme remarks: w=1/(σ2(F)+0.0001F2) [ Help ]
Δ/σ(max): 0.0449 [ Help ]
Δ/σ(mean): 0.0092 [ Help ]
Δρ(max): 6.78 e_Å-3 [ Help ]
Δρ(min): -8.13 e_Å-3 [ Help ]
Extinction method: none [ Help ]
Structural Information
Average Structure: (Show/hide table) [ Help ]
| Atom site label | Atom symbol | x | y | z | ADP type | Uiso/equiv | Symmetry multiplicity | Occupancy | Coords from (d)iffraction or (c)alculated | Coords restraints or constraints | Disordered cluster | Disordered group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bi | Bi | 0.5 | 0.15510(3) | 0.5 | Uani | 0.02038(19) | 4 | 1 | d | . | . | . |
| Br | Br | 0 | 0 | 0 | Uani | 0.0221(7) | 2 | 0.5 | d | . | . | . |
| B | B | 0 | 0 | 0.119175 | Uani | 0.0221(7) | 4 | 0.25 | d | . | . | . |
| O1 | O | 0 | 0.2440(11) | 0.5 | Uani | 0.014(3) | 4 | 1 | d | . | . | . |
| O2 | O | 0 | 0.104249 | 0.063088 | Uiso | 0.057(8) | 8 | 0.125 | d | . | . | . |
| O3 | O | 0.251554 | -0.060889 | 0 | Uiso | 0.057(8) | 8 | 0.125 | d | . | . | . |
| O4 | O | 0 | 0 | 0.42683 | Uiso | 0.1158 | 4 | 0.25 | d | . | . | . |
ADP components: (Show/hide table) [ Help ]
| Atom site label | Atom site symbol | U11 | U22 | U33 | U12 | U13 | U23 |
|---|---|---|---|---|---|---|---|
| Bi | Bi | 0.0291(4) | 0.0172(2) | 0.0148(3) | 0 | 0 | 0 |
| Br | Br | 0.0125(12) | 0.0214(10) | 0.0323(14) | 0 | 0 | 0 |
| B | B | 0.0125(12) | 0.0214(10) | 0.0323(14) | 0 | 0 | 0 |
| O1 | O | 0.012(5) | 0.019(4) | 0.013(5) | 0 | 0 | 0 |
Fourier Wave Vectors (explicit: q_x,q_y,q_z or coefficients: q_1,q_2,...): (Show/hide table) [ Help ]
| Wave vector code | q_1 |
|---|---|
| 1 | 1 |
| 2 | 2 |
| 3 | 3 |
Occupation crenel coefficients: (Show/hide table) [ Help ]
| Atom site label | Center (x0) | Width |
|---|---|---|
| Br | 0 | 0.5 |
| B | 0.5293 | 0.5 |
| O2 | 0.5155 | 0.5 |
| O3 | 0.5 | 0.5 |
| O4 | 0.6049 | 0.5 |
Definition of the displacive (translational) Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Displacement axis | Wave vector code |
|---|---|---|---|
| Bix1 | Bi | x | 1 |
| Biy1 | Bi | y | 1 |
| Biz1 | Bi | z | 1 |
| Bix2 | Bi | x | 2 |
| Biy2 | Bi | y | 2 |
| Biz2 | Bi | z | 2 |
| Bix3 | Bi | x | 3 |
| Biy3 | Bi | y | 3 |
| Biz3 | Bi | z | 3 |
| O1x1 | O1 | x | 1 |
| O1y1 | O1 | y | 1 |
| O1z1 | O1 | z | 1 |
| O1x2 | O1 | x | 2 |
| O1y2 | O1 | y | 2 |
| O1z2 | O1 | z | 2 |
Displacive (translational) Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| Bix1 | 0 | 0 |
| Biy1 | 0.00624(6) | 0 |
| Biz1 | 0 | 0.0458(4) |
| Bix2 | 0 | 0 |
| Biy2 | 0.00002(6) | 0 |
| Biz2 | 0 | -0.00719(18) |
| Bix3 | 0 | 0 |
| Biy3 | 0.00064(6) | 0 |
| Biz3 | 0 | 0.0082(2) |
| O1x1 | 0 | 0 |
| O1y1 | -0.0032(7) | 0 |
| O1z1 | 0 | -0.039(2) |
| O1x2 | 0 | 0 |
| O1y2 | -0.0067(10) | 0 |
| O1z2 | 0 | -0.013(3) |
Definition of the ADP Fourier series: (Show/hide table) [ Help ]
| Modulation code | Atom site label | Tensor element | Wave vector code |
|---|---|---|---|
| BiU111 | Bi | U11 | 1 |
| BiU221 | Bi | U22 | 1 |
| BiU331 | Bi | U33 | 1 |
| BiU121 | Bi | U12 | 1 |
| BiU131 | Bi | U13 | 1 |
| BiU231 | Bi | U23 | 1 |
| BiU112 | Bi | U11 | 2 |
| BiU222 | Bi | U22 | 2 |
| BiU332 | Bi | U33 | 2 |
| BiU122 | Bi | U12 | 2 |
| BiU132 | Bi | U13 | 2 |
| BiU232 | Bi | U23 | 2 |
ADP Fourier coefficients: (Show/hide table) [ Help ]
| Modulation code | Cosine coefficient | Sine coefficient |
|---|---|---|
| BiU111 | -0.0139(3) | 0 |
| BiU221 | -0.0098(2) | 0 |
| BiU331 | -0.0077(2) | 0 |
| BiU121 | 0 | 0 |
| BiU131 | 0 | 0 |
| BiU231 | 0 | -0.00517(19) |
| BiU112 | -0.0069(4) | 0 |
| BiU222 | 0.0019(4) | 0 |
| BiU332 | -0.0060(3) | 0 |
| BiU122 | 0 | 0 |
| BiU132 | 0 | 0 |
| BiU232 | 0 | -0.0016(2) |